CANDID-CNS™: Advancing CNS Penetration Prediction with Attentive Graph Neural Networks
4 min read
June 9, 2025
Publication
External Writer
Jesse W. Collins, PhD
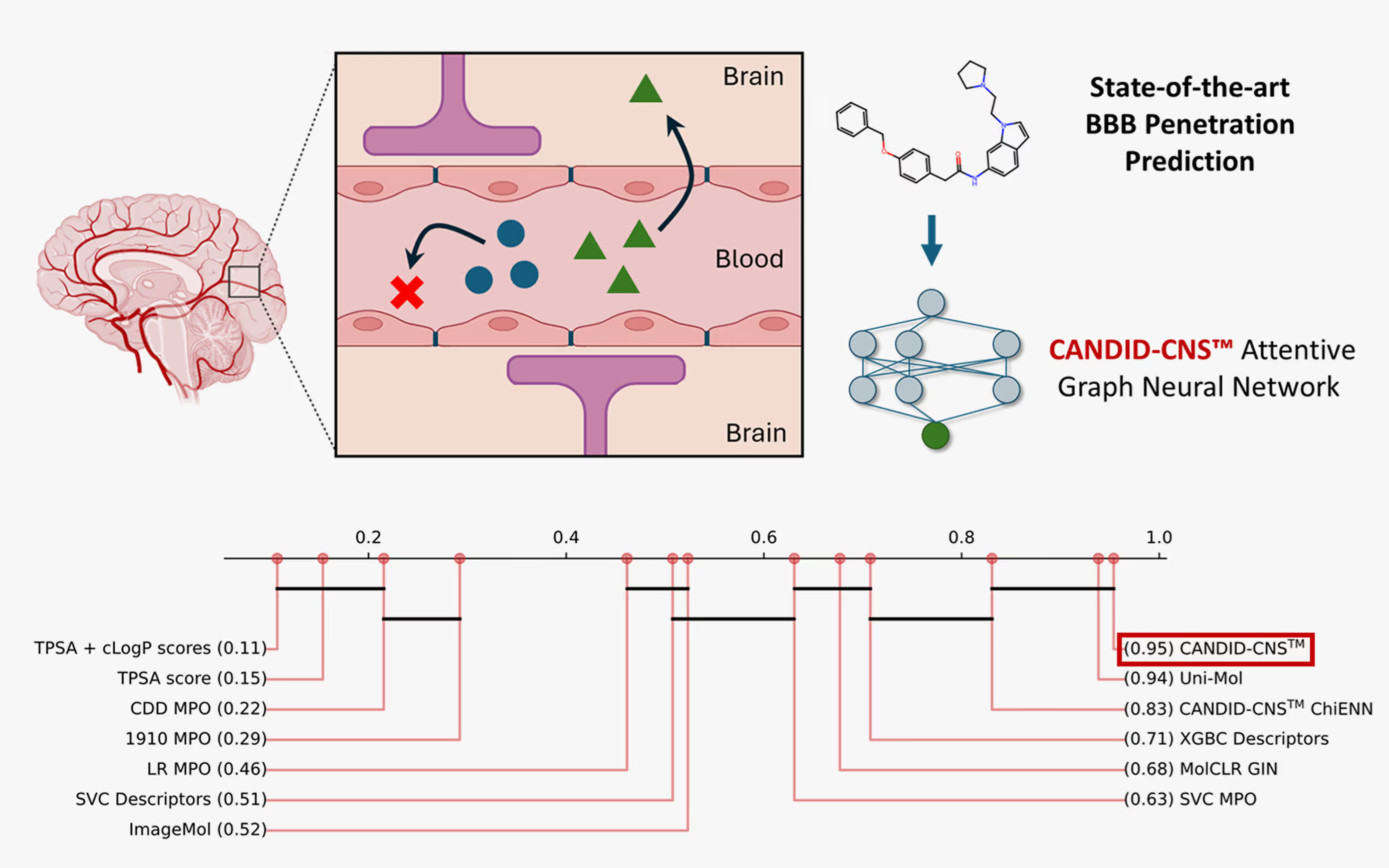
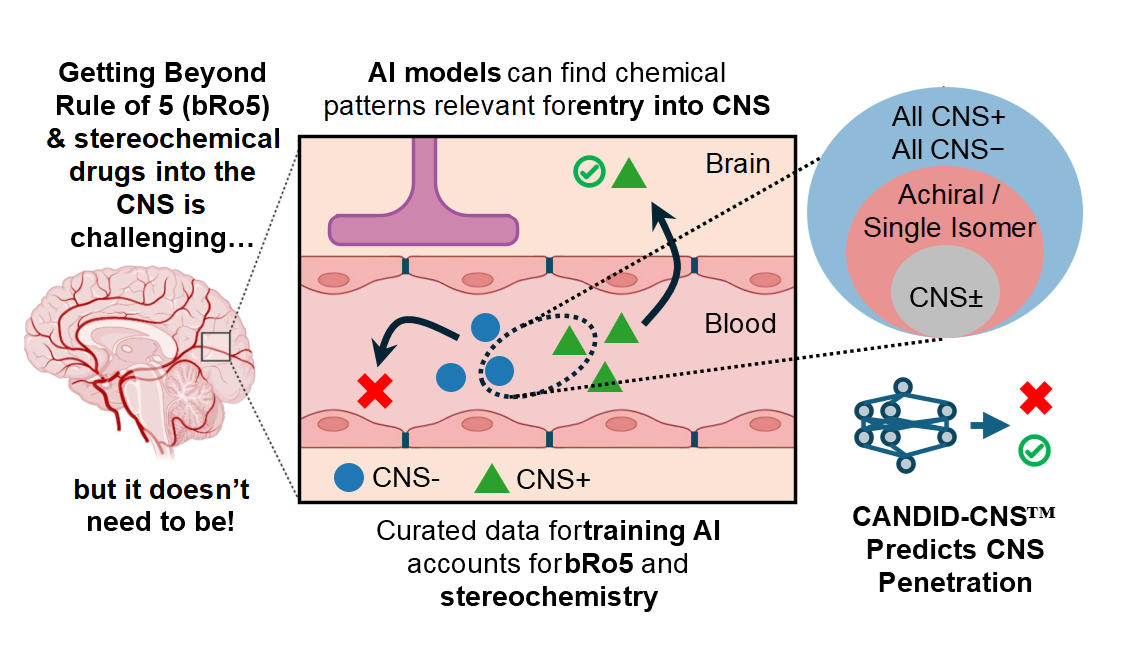
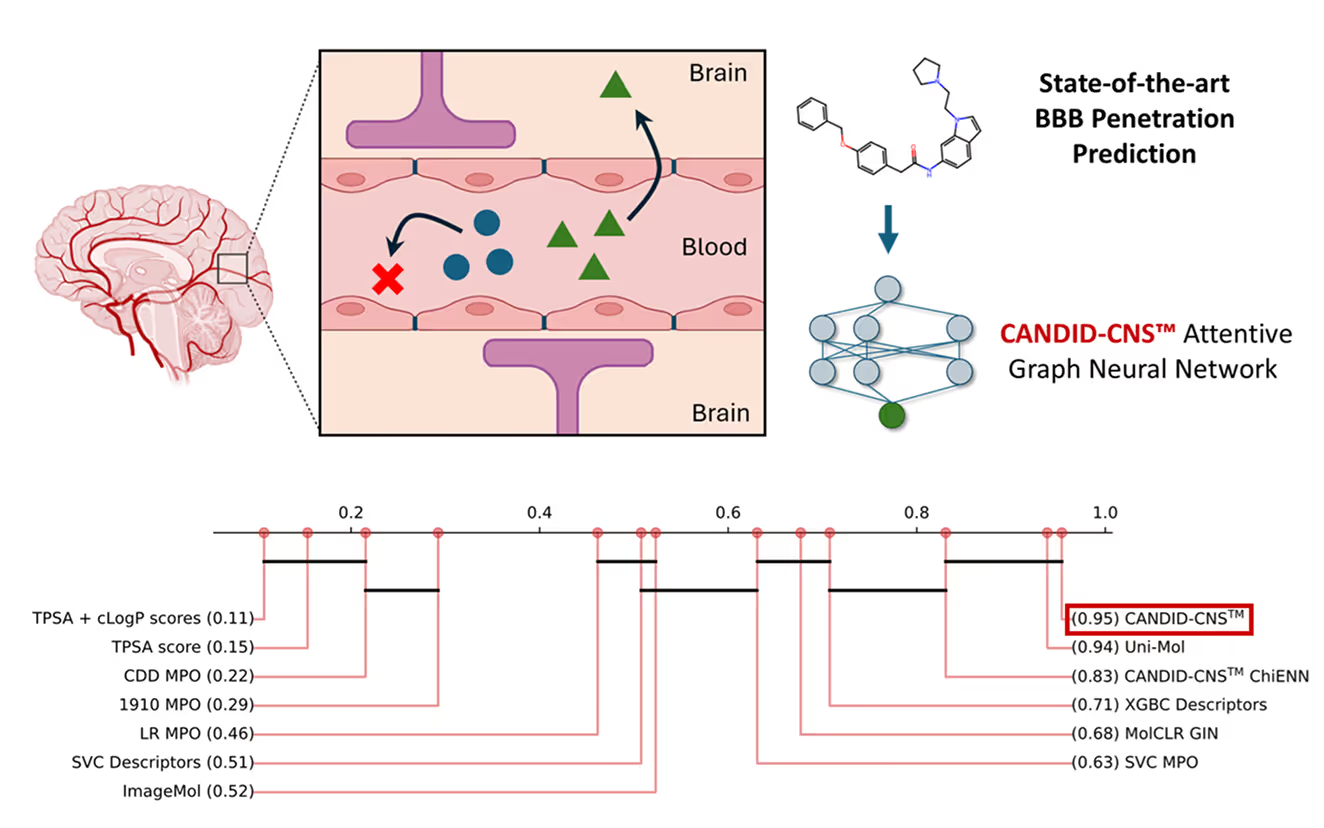
CANDID-CNS™ is 1910’s attentive graph neural network demonstrating best in class performance for predicting central nervous system (CNS) penetration, an important but difficult endpoint for neuroscience therapeutics to meet to become FDA-approved drugs. To treat conditions from the neurodegenerative, such as Alzheimer’s and ALS, to the neuropsychiatric, to glioblastomas and other cancers of the brain, small molecules must cross the blood-brain barrier (BBB), a protective layer of endothelial cells that carefully regulates human neurochemistry and prevents harmful chemicals and pathogens from damaging neuronal cells.
We compared CANDID-CNS™ to a variety of state-of-the-art neural networks and the industry standard CNS Multiparameter Optimization Score (CNS MPO). CANDID-CNS™ outperformed these on strict test sets that corrected for molecules known as “stereoisomers”, which have the same 2D chemical graph but different 3D structure. Most stereoisomers in the B3DB dataset we studied have the same CNS penetration class (BBB+ or BBB-), making it important to correct for this phenomenon when evaluating different models of CNS penetration.
Some groups of stereoisomers differ in CNS penetration class, however, and we refer to these as “multiclass stereoisomers.” We test whether CANDID-CNS™ and other approaches, including Uni-Mol, another top-performing model, which predicts CNS penetration based on 3D coordinate inputs, and which has 50 times as many adjustable parameters as CANDID-CNS™, could distinguish multiclass stereoisomers with statistical significance. Distinguishing between two stereoisomers that differ in CNS penetration is very difficult, and we did not expect any model to succeed better than random on these sets of molecules.
Surprisingly, CANDID-CNS™ distinguished multiclass stereoisomers better than random. The only models in our cross-validation study that succeeded on these most difficult multiclass stereoisomers were CANDID-CNS™ and a variant with a neural network layer specializing on chiral centers, called CANDID-CNS™ ChiENN [1].
This finding led us to investigate whether CANDID-CNS™ could perform well on larger molecules, which can have even more complex 3D configurations. We visualized how CANDID-CNS™ attended to the bonds of molecules it identified correctly as BBB+ that CNS did not. We observed that CANDID-CNS™ paid high attention to regions of molecules that were distant on 2D graphs, but appeared as if they might interact in 3D space, and to intermediate bonds with flexibility that could enable such “folding” and intramolecular interactions.
We performed quantum mechanical (QM) simulations and discovered that CANDID-CNS™ indeed identified bonds enabling complex 3D structures relevant to CNS penetration. Moreover, CANDID-CNS™ appears to have identified a broad pattern and design principle for CNS drug discovery. Many of the larger and more hydrophobic molecules CANDID-CNS™ correctly predicted to penetrate the CNS collapse in polar solvents such as water, forming intramolecular bonds e.g., via pi-pi stacking. In non-polar solvents, such as the cell membrane, the molecules expand. This solvent-dependent conformation change appears to enable surprisingly large and hydrophobic compounds to cross the BBB.
This pattern could influence medicinal chemists and AI-aided drug discovery alike in finding novel neuroscience therapeutics. CANDID-CNS™ and models like it could accelerate the lead optimization phase of drug discovery more broadly.
Preprint link:
Attentive graph neural network models for the prediction of blood-brain barrier permeability
References
[1] Gaiński, P.; Koziarski, M.; Tabor, J.; Śmieja, M. ChiENN: Embracing Molecular Chirality with Graph Neural Networks. arXiv July 10, 2023. https://doi.org/10.48550/arXiv.2307.02198.